This post is meant for people who are new to deep learning and are looking for ways to quickly build neural network models. They may not necessarily be from a math or computer science background, but have experience with python and interested in using deep learning in your own domain. Its important to know that in real life, there is no free cookies and deep learning is no exception. Yes, the basics of how it works is very simple, but to truly understand deep learning, one has to dig deeper and go beyond the basics. It may be easy to learn but its really difficult to master. The bottom line is that even though most online courses or blogposts (including this one) may give you the impression that deep learning is quiet fun and easy, it is only meant to spark interest, but one should always remember that at its core deep learning is all about one thing, maths.
 Nonetheless, We all need to start somewhere, so we might as well begin by making use of frameworks like fastai, to help us implement neural networks quickly, so we can visualise what’s happening and get an intuitive sense of how deep learning could help us in our own domain.
Nonetheless, We all need to start somewhere, so we might as well begin by making use of frameworks like fastai, to help us implement neural networks quickly, so we can visualise what’s happening and get an intuitive sense of how deep learning could help us in our own domain.
Defining the problem being solved:
Since I come from a biomedical background, I am obviously more interested in understanding how deep learning could impact the future of medical research and treatment. When most people think of the future of technology in medicine, they imagine robots performing surgery or a breakthrough in treatment regime, but in reality what’s most needed is an innovation in the field of diagnosis especially in pathology. Even today most pathologists still run glass slides with tissue or fluid samples from lab to lab for consultation, and rely heavily on manual slide viewing through an optical microscope. This is really inefficient and prone to errors. Automated cell classification is thus an important yet a challenging computer vision task with significant benefits to biomedicine.
In this blog, we’ll attempt to use the fastai library (thanks to Jeremy Howard and his team at USF) to build an image classifier that works amazingly well for a classification task. We will try to replicate the work of Yang Yujiu, et. al., where they proposed a convolutional neural network (CNN) based method to classify human breast-derived cell lines based on actin filament organisation. The cells they wanted to classify were as follows:
- normal breast epithelial cell line, MCF-10A (non-aggressive)
- breast cancer cell lines, MCF-7 (less aggressive)
- MDA-MB-231 (more aggressive)
They collected a total of 552 images were captured, in which each image contained at least three individual cells, resulting in more than 1500 individual cell images (approximately 500–600 individual cell images for each type of cell). To be specific, MCF-10A, MCF-7 and MDA-MB-231 classes each contained 182, 186 and 184 images, respectively. The goal of this work is to train a convolutional neural network on the their dataset and achieve close to, or near state-of-the-art results.
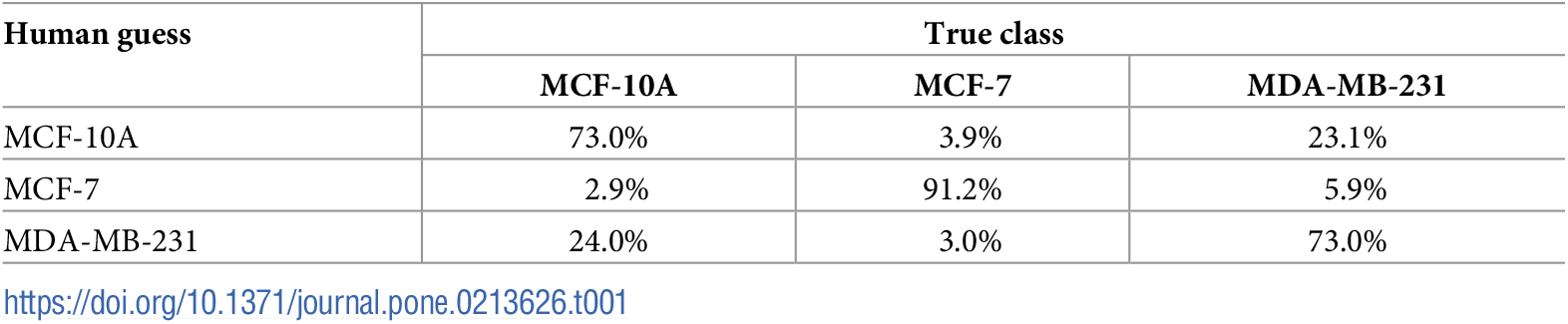
|
|---|
| Shows the confusion matrix of cell classification task performed by human expert |
| This blogposts presents research and an analysis of their dataset using Fastai + Pytorch and is provided as a reference, tutorial, and open source resource for others to refer to. It is not intended to be a production ready resource for serious clinical application. We work here instead with low resolution versions of the original high-res images for education and research. This proves useful ground to prototype and test the effectiveness of various deep learning algorithms. |
For this blog post we will approach the problem in 3 steps:
- loading data into the right format
- creating an instance of our neural network model and training on our data
- learning how to create final adjustments and tweaking to make the model better
Loading our Data:
Before we can begin to build our model it’s important that we explore, prepare and load our dataset into a format ready for learning model to work with. Fastai comes with an amazing data_block api, which allows us to load data from different sources. For this post I will be using pictures that are downloaded and kept in different folders. The name of the folder acts as label value for our images, which we specify in a variable called classes. The code below shows how we can load the images into a format that is ready for our CNN model to work with.
path = Path('/storage/data') #specify the path containing the folder
classes = ['MDA-MB-231', 'MCF-10A', 'MCF-7'] #these act as our labels for images
np.random.seed(42)
# then we load the data using the ImageDataBunch class
data = ImageDataBunch.from_folder(path, train=".", valid_pct=0.2, ds_tfms=get_transform(), size= 480, num_workers=4).normalize(imagenet_stats)
data.show_batch(3,figsize=(12,12), hide_axis= True)
We can then check how our images look like
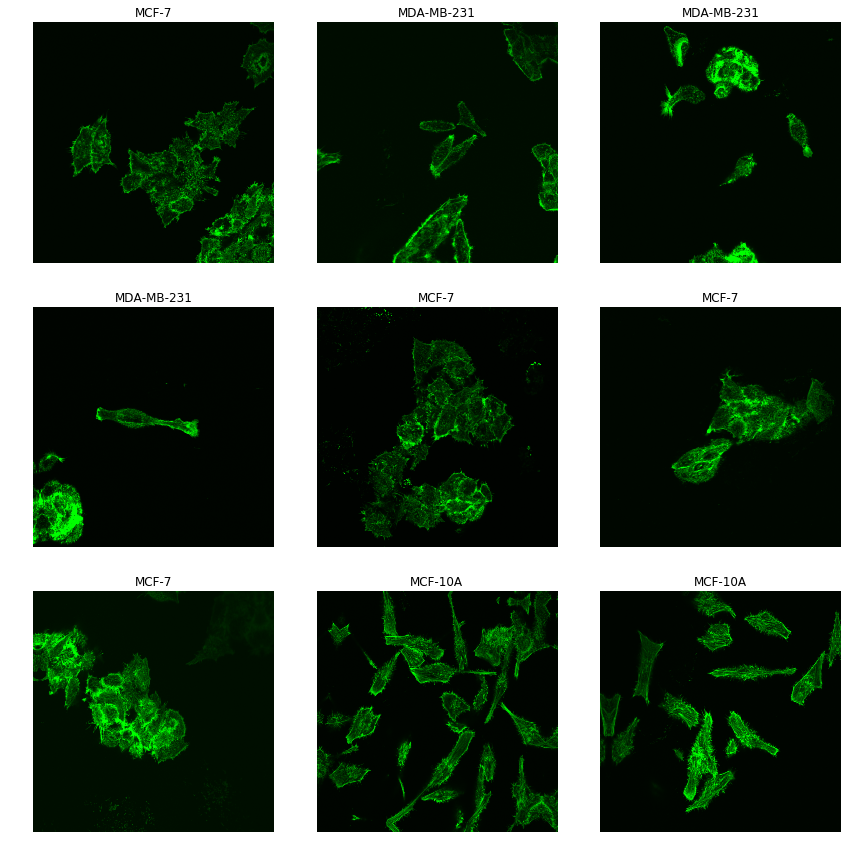
Creating our model
Once we have a correctly setup the ImageDataBunch object, we can now pass this, along with a pre-trained ImageNet model, to a cnn_learner. The following line instantiates the model that we’ll use to perform classifications with. The Resnet model we use will be a pre-trained model on the imagenet database.
learn = cnn_learner(data, models.resnet34, metrics=error_rate)
In the following line, we actually go ahead and fit the model. The way fastai library is organised we usually just need to decide between 3 things to build the most simplest model.
- The model architecture (in our case we use resnet34)
- epoch (number of times we want to pass our dataset through the network)
- Learning rate (tuning parameter that determines the step size)
learn.fit_one_cycle(3)
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix(figsize=(6,4), dpi=100)
We can begin with 3 epoch cycles and see how well our model performs, we get the following output

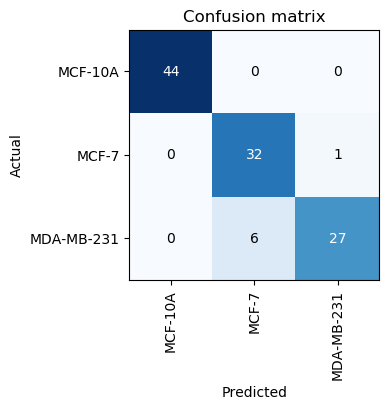
With just 3 cycles we manage to get an error of less than 7%, which is already better than performance of human experts. We can check the confusion matrix to learn more. Our model is good at identifying the normal cell line but is still not good and classifying MCF-7 and MDA-MD-231.
Fine tuning our parameters:
Even though we have a pretty good model, we can do better. Usually for people from non-computer science background, this tends to be the part we get to actually shine by using our domain expertise. First thing we do is obtain the data images which were predicted wrongly and look at the probability values associated with each images. By looking at the images we can then try and understand what went wrong, and then try a number of corrective approaches. Please take a look at the fastai docs to learn more. Using fastai library we can get these wrongly classified images in just 1 line of code.
interp.plot_top_losses(9, figsize=(12,12))
When we look at the images of misclassified cells we do not see any noticeable pattern. When cells are aggregated together they may cause our model to misclassify them, but thats not true at-least in our case as I don’t see any obvious pattern.
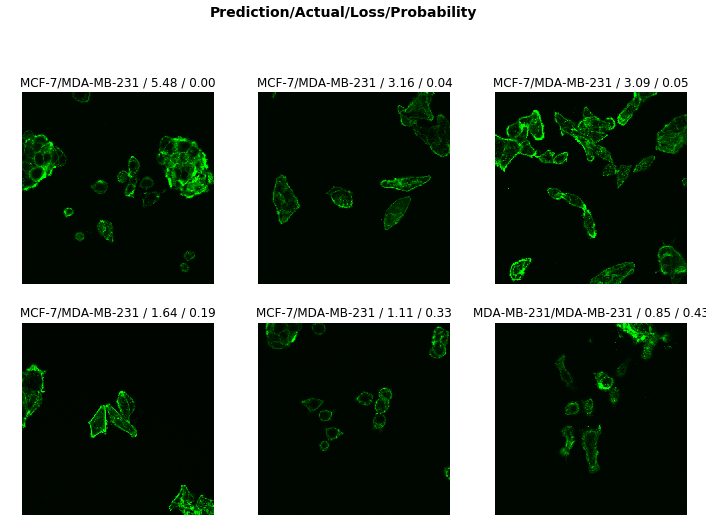
We can try to rotate our images or carry out some image enhancement or data augmentation methods but it doesn’t seem necessary. We can instead try to plot our error rate against learning rate. For neural networks, the learning rate is basically how quickly a network abandons old beliefs for new ones.
If a child sees 10 examples of cats and all of them have orange fur, she will think that cats have orange fur and will look for orange fur when trying to identify a cat. Now she sees a black cat and her parents tell her it’s a cat (supervised learning). With a large “learning rate”, the child will quickly realise that “orange fur” is not the most important feature of cats. With a small learning rate, she will think that this black cat is an outlier and cats are still orange.
So in short, choosing the right learning rate can help train our model to spot outliers. We can do this using fastai library in just a few lines of code and obtain the learning rate.
learn.unfreeze()
learn.fit_one_cycle(1)
learn.lr_find()
learn.recorder.plot()
We train our model for just 1 cycle so we can see how the error rate is change in one cycle.
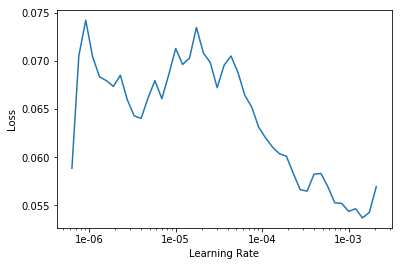
We can see that our loss function shows a sharp decrease between learning rate of 1e-5 to 1e-3. This means that if we train our model keeping the learning rate between these learning rate it might improve its performance.
So we fit the model once again but this time setting new parameter for learning rate. We do that as follows
learn.unfreeze()
learn.fit_one_cycle(2, max_lr=slice(1e-5,1e-3))
We just need to specify another parameter in our function called max_lr and put the value of learning rate where we noticed the sharpest decay.
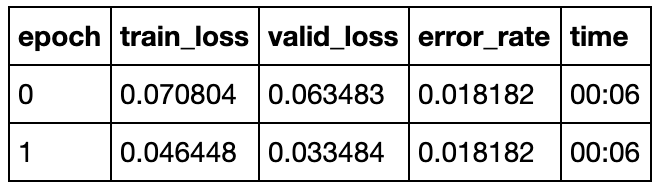
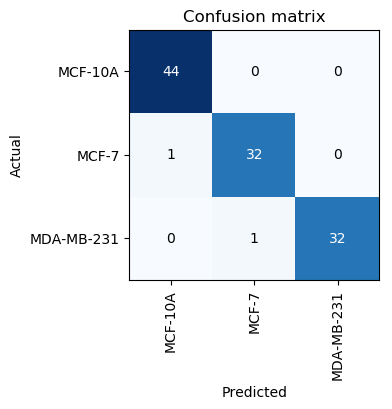
So this time, with just two cycles we have managed to get accuracy of around 98.2%. Our model can capture the difference effectively. The only difference is that this time it managed to miss one aggressive cell image and classified it as the normal MCF-10A. So there is still room for improvement.
References:
Oei RW, Hou G, Liu F, Zhong J, Zhang J, An Z, et al. (2019) Convolutional neural network for cell classification using microscope images of intracellular actin networks. PLoS ONE 14(3): e0213626. https://doi.org/10.1371/journal. pone.0213626
fast.ai — Online course on Practical DeepLearning for Coders